Results
Variant Mutation Assay Specificity and Sensitivity
In silico analysis indicated no cross-reactivity between the assays and deposited sequences in GenBank. When challenged against wild-type gRNA, the respiratory virus panel, and actual or synthetic variant gRNA, no cross-reactivity occurred. Positive controls and no-template controls run on the sample plate performed according to expectations. Variant mutation concentrations were measured in no, low, and high background of wild-type gRNA, which does not contain the mutations. Results of mutation assays in the presence of high and low background wild-type gRNA are similar to their results in the absence of background wild-type gRNA (Appendix Figure 1), indicating that the assays are sensitive and specific.
Variant Mutation Concentrations in Wastewater Solids
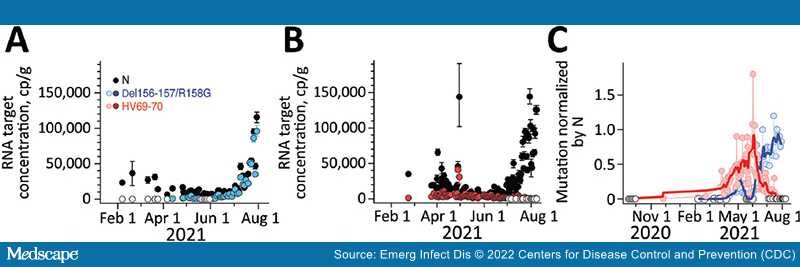
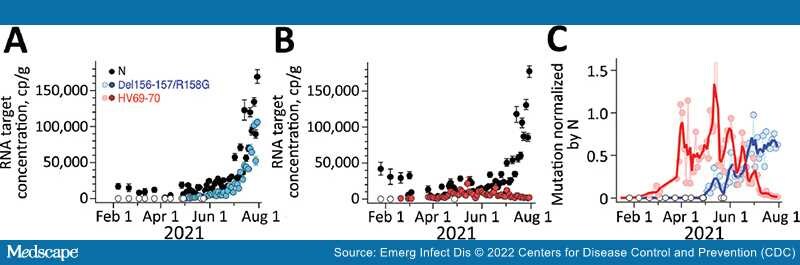
Results for positive and negative controls were as expected, and recovery controls indicated consistent RNA recovery from samples and lack of substantial inhibition (Appendix). We measured HV69–70 concentrations up to 1 time/day at San Jose and up to 3 times/week at Sacramento; concentrations ranged from not detected to >10,000 copies/g (Figure 1, 2). N and HV69–70 concentrations at San Jose before 15 Feb 2021 are not presented graphically; samples collected during July and September 2020 did not have measurable HV69–70. HV69–70 was measured for the first time in San Jose solids on November 25, 2020, at concentrations of ≈103 copy/g. We did not detect HV69–70 in Sacramento wastewater solids before late February 2021; results for samples collected in October 2020 (not shown in plot) and late January 2021 were nondetect for HV69–70. At both locations, the concentration of HV69–70 relative to the N gene (HV69–70/N ratio) increased over time beginning in early March 2021, peaked in early June 2021 at San Jose and May 2021 at Sacramento when HV69–70/N was ≈1, and then fell until HV69–70 became undetectable at San Jose and present at very low relative concentrations at Sacramento (0.01) in late July 2021 (Figure 1, 2).
Figure 1.
Measurements of severe acute respiratory syndrome coronavirus 2 variants of concern in wastewater solids, San Jose, California, USA. Concentrations of N gene and mutations found in Delta (Del156–157/R158G; panel A), and Alpha (HV69–70; panel B) variants in wastewater solids and their ratio (panel C). Error bars in panels A and B represent SDs derived from the 10 replicates run for each sample; open white circles are nondetects (below the limit of detection) and shown as 0. Errors include technical and replication errors. If error bars are not visible, then errors are smaller than the symbol. Panel C shows smoothed lines for visual reference for mutation ratios. For Del156–157/R158G/N ratio, the smoothed line is a 3-point running average, and for the HV69–70/N ratio, the smoothed line is a 7-point running average; each approximates a weekly average. The timescale for the HV69–70 data (B) is truncated for visualization; additional data on dates before February 15, 2021, are described in the article and shown (C), with the exception of data from July 14, 2020, which was nondetect. N, nucleoprotein.
Figure 2.
Measurements of severe acute respiratory syndrome coronavirus 2 variants of concern in wastewater solids, Sacramento, California, USA. Concentrations of N gene and mutations found in Delta (Del156–157/R158G; panel A), and Alpha (HV69–70; panel B) severe acute respiratory syndrome coronavirus 2 in wastewater solids and their ratio (panel C). Error bars in panels A and B represent SDs derived from the 10 replicates run for each sample; open white circles are nondetects ((below the limit of detection) and shown as 0. Errors include technical and replication errors. If error bars are not visible, then errors are smaller than the symbol. For Del156–157/R158G/N ratio, the smoothed line is a 3-point running average, and for the HV69–70/N ratio, the smoothed line is a 7-point running average; each approximates a weekly average. The timescale for the HV69–70 data (B) is truncated for visualization; additional data on dates before January 15, 2021, are described in the article and were nondetects. One data point is located beyond the upper bound of the y-axis (C): the value for HV69–70/N on May 14, 2021, was 2.4. N, nucleoprotein.
Del156–157/R158G concentrations were measured as frequently as three times per week at both San Jose and Sacramento and ranged from not detected to 100,000 copies/g (Figure 1, 2). We observed Del156–157/R158G nondetects in samples collected before early April 2021 at both sites, and then both sites experienced a small peak in Del156–157/R158G concentration in early to mid-May 2021 (Del156–157/R158G relative to N ≈0.2–0.3 at the 2 sites), followed by a decline to undetectable levels over ≈2 weeks, followed by a sharp increase until the end of the data series. During this time, N gene concentrations in wastewater increased contemporaneously. The concentration of Del156–157/R158G relative to N (Del156–157/R158G/N ratio) (Figure 1, 2) increased to ≈0.8 at the sites by the end of the data series.
Trends in Variants in Sequenced Case Isolates From Sewersheds
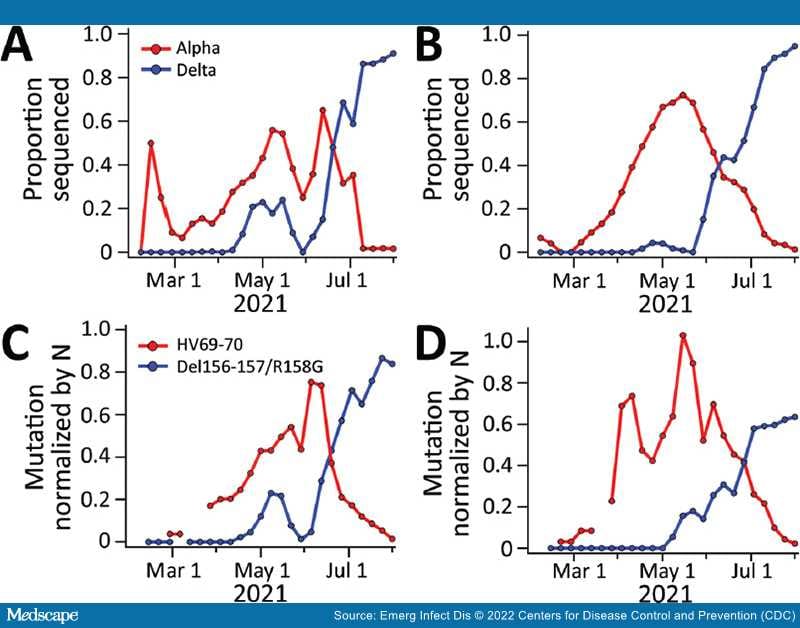
We analyzed trends in Alpha and Delta variants confirmed from case isolates collected from residents (case isolates) of the San Jose and Sacramento sewersheds from early February through late July 2021 (Figure 3). Alpha proportions increased in both sewersheds from early March, peaking in May–June and decreasing in early July. Delta was first identified in isolates in early April and by the end of July accounted for almost all sequenced isolates. In San Jose, a small peak in Delta occurred in May, before a large sustained increase in June; a similar peak is also evident, to a lesser extent, in the Sacramento Delta data. During this period, the 7-day average laboratory-confirmed incident COVID-19 cases ranged from 1 to 30/100,000 population (Appendix Figure 2) in each sewershed. Incident COVID-19 cases in each sewershed is positively and significantly correlated with N gene measurements in the settled solids (Pearson R [rp] 0.8, df 46–131, p<10–10 for both San Jose and Sacramento N gene datasets, regardless of whether they were generated when measuring the Delta or Alpha mutation).
Figure 3.
Comparison of severe acute respiratory syndrome coronavirus 2 variants of concern in wastewater solids with coronavirus disease case isolates, San Jose and Sacramento, California, USA, February 1–July 31, 2021. A, B) Proportion of circulating severe acute respiratory syndrome coronavirus 2 attributable to the Alpha and Delta variants, estimated from isolate sequencing data from cases collected and sequenced over the previous 14-day period in San Jose (A) and Sacramento (B) sewersheds. C, D) Concentrations of mutations found in Alpha (HV69–70) and Delta (Del156–157/R158G) variant viruses, normalized by N gene concentrations in wastewater, averaged over the previous 14 days in San Jose (C) and Sacramento (D). No data are shown for dates for which no measurements were made within the previous 14 days. N, nucleoprotein.
Relationship Between Proportion of Alpha and Delta Variants in Case Isolates and Wastewater Mutation Data
We compared ratios of HV69–70 and Del156–157/R158G mutations to the N gene (VOC abundance estimates based on wastewater) against the proportion of all case isolates sequenced and identified as Alpha and Delta variant (VOC abundance estimates based on case isolate sequencing) from each sewershed (Figure 3). Trends of wastewater VOC abundance estimates follow closely and temporally the trends of case isolate sequencing VOC abundance estimates during this period at both sewersheds, including features such as an early peak in Delta in May. Alpha and Delta mutation gene ratios from wastewater were strongly correlated with the corresponding ratios of each VOC from case isolates sequenced: rp 0.82 (p<10–5, df 19 in San Jose) and 0.88 (p<10–7, df 21 in Sacramento) for Alpha; rp 0.97 (p<10–15, df 23 for both San Jose and Sacramento) for Delta. When compared to the opposing variant, the mutation gene ratios were not correlated (p>0.05 for all).
Completeness of and Delays in Receiving SARS-CoV-2 Isolate Sequence Data
During February 1–August 1, 2021, the total number of case isolates sequenced over a 14-day period in our 2 sewersheds varied (2–520 median for 8% of all sequenced case isolates from San Jose and 6% from Sacramento). Earliest isolate sequencing results were available to CDPH ≈5 days after sample collection date. Approximately 75% of all sequenced isolate results in our dataset were available within 2–3 weeks. As more isolate sequencing data were received, estimated proportions of VOCs changed over time. Around 3 weeks were required for 95% of VOC estimates (14-day window) to be within 10% of the final estimate.
Emerging Infectious Diseases. 2022;28(5):940-947. © 2022 Centers for Disease Control and Prevention (CDC)