Abstract and Introduction
Introduction
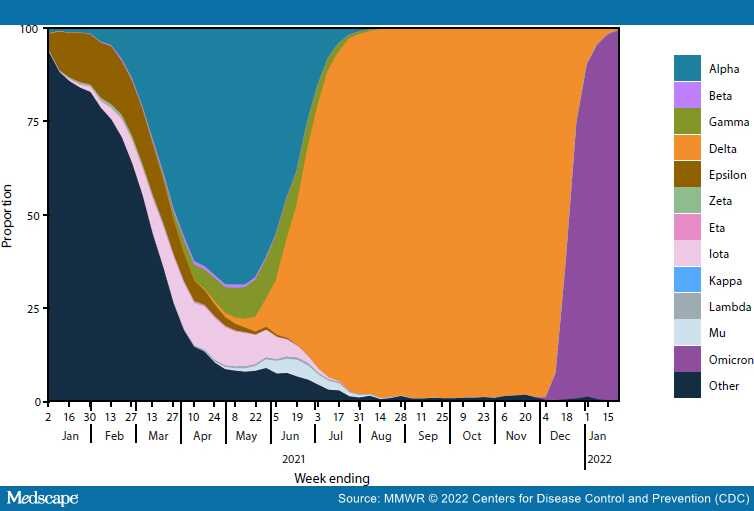
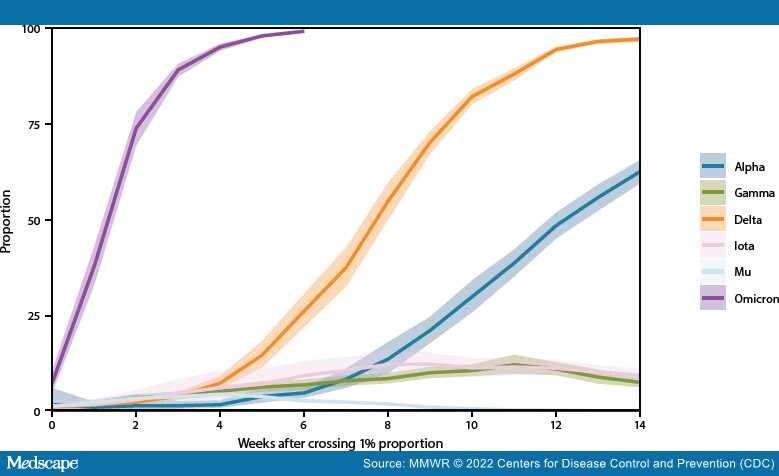
Genomic surveillance is a critical tool for tracking emerging variants of SARS-CoV-2 (the virus that causes COVID-19), which can exhibit characteristics that potentially affect public health and clinical interventions, including increased transmissibility, illness severity, and capacity for immune escape. During June 2021–January 2022, CDC expanded genomic surveillance data sources to incorporate sequence data from public repositories to produce weighted estimates of variant proportions at the jurisdiction level and refined analytic methods to enhance the timeliness and accuracy of national and regional variant proportion estimates. These changes also allowed for more comprehensive variant proportion estimation at the jurisdictional level (i.e., U.S. state, district, territory, and freely associated state). The data in this report are a summary of findings of recent proportions of circulating variants that are updated weekly on CDC's COVID Data Tracker website to enable timely public health action.†The SARS-CoV-2 Delta (B.1.617.2 and AY sublineages) variant rose from 1% to >50% of viral lineages circulating nationally during 8 weeks, from May 1–June 26, 2021. Delta-associated infections remained predominant until being rapidly overtaken by infections associated with the Omicron (B.1.1.529 and BA sublineages) variant in December 2021, when Omicron increased from 1% to >50% of circulating viral lineages during a 2-week period. As of the week ending January 22, 2022, Omicron was estimated to account for 99.2% (95% CI = 99.0%–99.5%) of SARS-CoV-2 infections nationwide, and Delta for 0.7% (95% CI = 0.5%–1.0%). The dynamic landscape of SARS-CoV-2 variants in 2021, including Delta- and Omicron-driven resurgences of SARS-CoV-2 transmission across the United States, underscores the importance of robust genomic surveillance efforts to inform public health planning and practice.