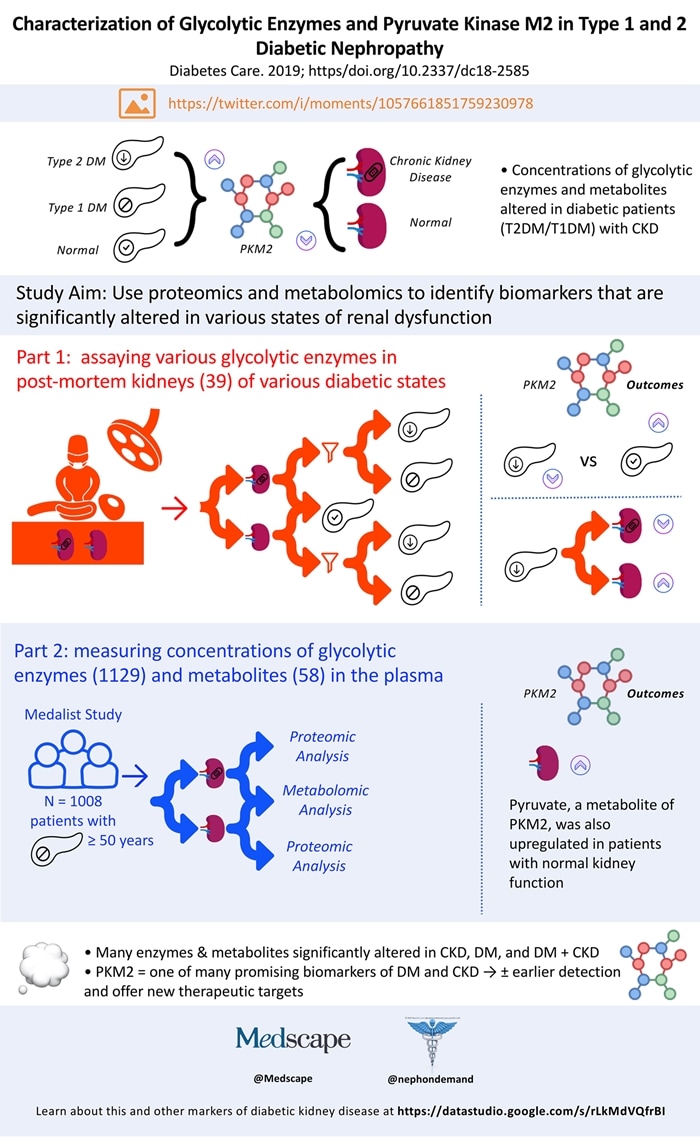
We live in an age of "big data." One could argue that nephrology was at the forefront of such analyses with early trials using USRDS and DOPPS data. Proteomic and metabolomic studies are representative of modern-day big-data analyses and distinct from databases such as USRDS and DOPPS. Traditional databases include data from a large number of patients over an increasing period of time, but proteomic/metabolomic studies analyze large volumes of data generated by a small number of patients in a shorter period of time. This feature makes such studies attractive for revealing patterns that may otherwise take years to uncover. One such study, by Gordin and colleagues , combines proteomic and metabolomic analyses to identify new targets for diabetic kidney disease.
They focused on glycolytic pathway enzymes (and their metabolites) and pyruvate kinase M2 (PKM2) in particular. PKM2 is upregulated in patients with diabetes who do not have chronic kidney disease(CKD). It and other upregulated enzymes are thought to play a role in protecting against diabetic kidney disease. The researchers took two approaches to identify PKM2 and other biomarkers that could provide kidney protection against diabetes. Part one entailed tissue sampling of post-mortem kidneys, and part two used proteomic and metabolomic analyses.